"Mom does it best: Parental Care as a Model Phenotype to Explore How Cell-Type Specific Changes in Gene Expression Influence Brain Activity and Animal Behavior"
 Dr. Brandon Logeman
Dr. Brandon Logeman
Bio:
Brandon L. Logeman, PhD is a new Assistant Professor in the Department of Molecular and Cellular Biochemistry, College of Medicine, University of Kentucky. After completing his Ph.D. at Duke University, he joined the lab of Catherine Dulac at Harvard University to study the molecular mechanisms through which changes in cell-type specific gene expression influence neural activity and animal behavior. After receiving a K99/R00 Career Transition Award he joined the University of Kentucky in August 2025. His new lab will utilize custom designed single-cell genomics technologies such as microfluidic, droplet based sequencing assays and imaging based spatial transcriptomics as well as de novo protein binder design across a panel of genetically diverse mouse strains to discover how genomic and environmental influences contribute to observable differences in animal behavior.
Abstract:
Parental care is composed of multiple infant-directed behaviors that promote offspring survival and is influenced by the sex and physiological state of the caregiver. Previous work in mice has identified the medial preoptic area of the hypothalamus as a key brain area implicated in parental behaviors. However, numerous naturalistic behaviors and homeostatic processes are controlled by this area, hindering mechanistic investigation of the circuits underlying parental care. To overcome this challenge, here I employ cell-type specific RNA- and ATAC-seq analysis, neural activity recording, and perturbation to gain access into molecular, biophysical, and circuit-based causality of behavioral control. I find that various neuronal types involved in parenting behavior are each distinctively influenced by the sex and physiological status of an individual and uncover how cell-type specific regulatory programs alter gene expression and neural activity underlying behavior control. These results demonstrate how cell-type specific transcriptional responses to internal physiological cues mediate circuit specific alterations to neural activity and ultimately influence animal behavior.
"Circadian Rhythms in Hosts, Parasites and their Vectors”
 Dr. Filipa Rijo-Ferreira | Rijo-Ferreira Lab
Dr. Filipa Rijo-Ferreira | Rijo-Ferreira Lab
Bio:
Filipa is an assistant professor of public health and molecular and cellular biology at the University of California, Berkeley, and an investigator at the Chan Zuckerberg BioHub. Filipa earned a Ph.D. from University of Porto, Portugal, followed by postdoctoral training at the University of Texas Southwestern Medical Center at Dallas. The Rijo-Ferreira lab takes an integrated approach to study circadian rhythms in parasitic diseases in particular Malaria and Sleeping sickness. Filipa is a NIH Pathway to Independence awardee, a NIH Director’s New Innovator awardee and a Searle Scholar. Moreover, Filipa was recently named a HHMI Freeman Hrabowski Scholar that recognizes outstanding early career faculty for their impactful research and commitment to mentoring.
"Hybridization and Adaptation in Chickadees"

Dr. Scott Taylor | Taylor Lab
Bio:
Dr. Scott Taylor is the director of the CU Boulder Mountain Research Station. Research in his group focuses on using natural hybrid zones and recent radiations to understand the genetic bases of traits involved in reproductive isolation, population divergence and speciatio, and the impacts of anthropogenic change, including climate change, on species distributions, interactions and evolution. We're fascinated by natural history and the intersections between art and science, and we're committed to doing our part to support our community.
Abstract:
Chickadees are familiar and widespread nonmigratory birds that have amazing adaptations to survive cold winters, including the ability to remember hundreds of thousands of individual seed cache locations. Recovering these caches is critical for winter survival. Taylor will share recent results from two different studies, one focused on a chickadee hybrid zone and one focused on a single population of mountain chickadees, that both provide insights into the genetic basis of variation in spatial cognition in chickadees.


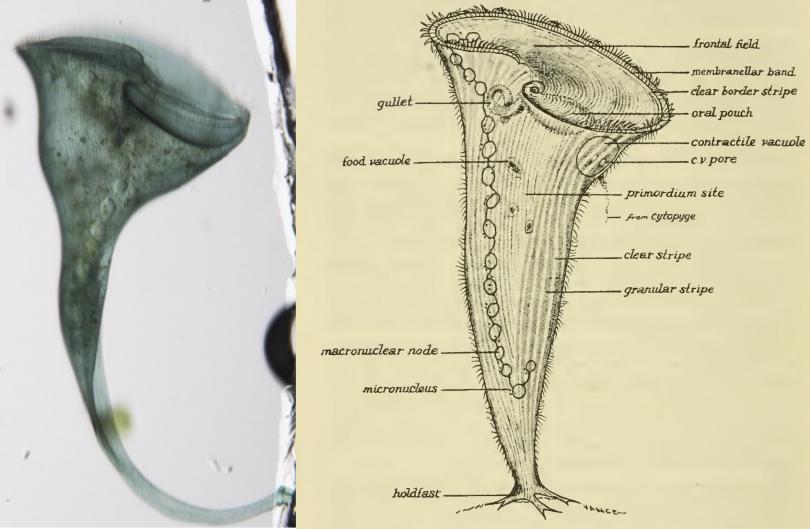
"Pattern Formation and Regeneration in a Single Cell"
 Dr. Wallace Marshall | Marshall Lab
Dr. Wallace Marshall | Marshall Lab
Bio:
Wallace Marshall is professor of biochemistry and biophysics at the University of California, San Francisco. A native Long Islander, he received his bachelor’s degrees in electrical engineering and biochemistry from the State University of New York at Stony Brook and his Ph.D. in biochemistry from UC San Francisco, where he studied organization of chromosomes within the nucleus with John Sedat. He then moved to Yale University for postdoctoral studies with Joel Rosenbaum, where he became interested in questions of organelle size control and cell organization, using cilia, flagella and centrioles as model systems.
In 2003, he joined the faculty at UCSF, where he continues to study questions of cellular organization in a variety of model organisms including green algae, yeast, ciliates and mammalian cells. In recent years, his research program has expanded to include cellular engineering and the study of learning and computation in single cells. Marshall is an elected fellow of the American Society for Cell Biology, former co‐director of the physiology summer course at the Marine Biological Laboratory in Woods Hole, Massachusetts, and he currently is director of the Center for Cellular Construction, an NSF Science and Technology Center, devoted to engineering cells and tissues.
Abstract:
Cells are sometimes viewed as small, simple building blocks for more complex organisms, but in fact individual cells can have extremely complex structures. Where does the information come from to build patterns within a single shared cytoplasm? We view this as essentially a question of developmental biology, but now at the sub-cellular level.
To understand how developmental processes work within a single cell, we have turned to the classical model organism Stentor coeruleus. Stentor is a giant ciliate with a size and complexity comparable to an animal embryo and has extraordinary powers of regeneration that classically allowed surgical manipulations similar to those used to for studying development in embryos. We have sequenced the Stentor genome and developed methods to perturb gene expression.
Our current work focuses on two questions:
- How does the cell sense when it is necessary to regenerate?
- How does the cell build and maintain patterns corresponding to the major body axes?
Using a combination of genomics, proteomics, and microscopy methods, we have begun to make progress in both directions. We find that the initiation of regeneration appears to involve a cell cycle transition governed by the Cdk4/cyclin D - E2F pathway as well as by MAPK signaling. Development of spatial pattern appears to involve non-random distribution of cytoskeletal scaffolding proteins and localization of mRNA encoding both structural proteins and potential transcription factors, possibly suggesting a role for localized differences in gene expression within the highly polyploid macronucleus.